Dickinson Strategies
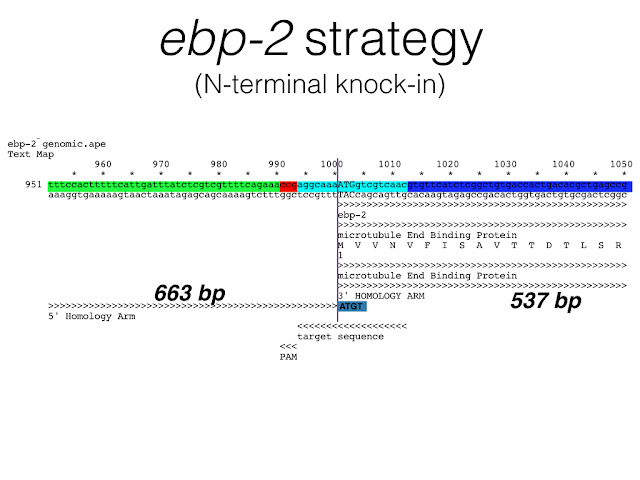
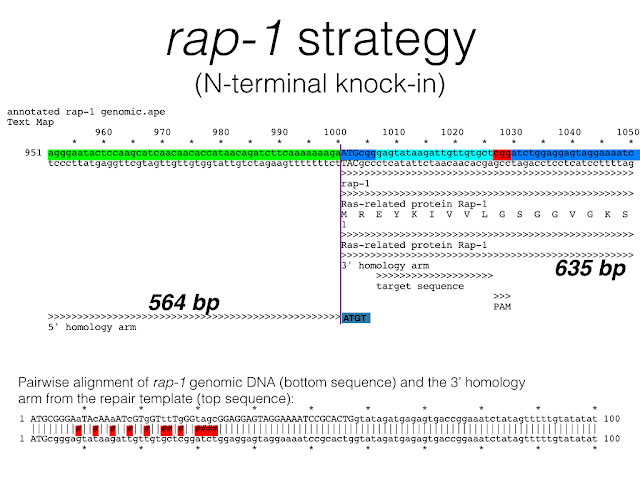
Dickinson et al. 2015 created fluorescent protein knock-ins for a number of genes in C. elegans using the genome editing tool, CRISPR. The following are three annotated text maps that include 100 bp of genomic DNA centered around the target sequence (aqua blue) and the PAM (red). The 3' end of the 5' homology arm and the 5' end of the 3' homology arm are positioned as indicated (The color scheme is a little misleading since it obscures the true ends of the homology arms in most cases). Focus instead on the text annotations below the sequence. In all cases, there is no gap between the end of the 5' homology arm and the beginning of the 3' homology arm used for repair. This is to ensure that no sequence is gained or lost during homologous recombination. Also notice that when the target sequence + PAM is positioned completely within a homology arm, silent mutations are incorporated into that homology arm to prevent cleavage of the repair template (see rap-1 and par-5). This is illustrated with a pairwise alignment between the genomic DNA sequence and the homology arm sequence inserted into the repair template construct.
Subscribe to:
Comments (Atom)

No comments:
Post a Comment
Note: Only a member of this blog may post a comment.